S
S
S
ANKARA UNIVERSITY COMPSEG
HISTOPATHOLOGY IMAGES RESEARCH GROUP
TUBITAK project of 121E379 titled “Development of Deep Learning based Methodology for the Detection of the Breast Cancer in Histopathology Images”
NuSeC and MiDeSeC Datasets
Definition of NuSeC and MiDeSeC Datasets
(NuSeC and MiDeSeC Datasets Description)
1.Introduction
The datasets of NuSeC (Nuclei Segmentation and Classification) and MiDeSeC (Mitosis Detection, Segmentation and Classification) were created and used in training and testing of deep learning models for breast cancer detection and grading within the scope of TUBITAK project of 121E379 titled “Development of Deep Learning based Methodology for the Detection of the Breast Cancer in Histopathology Images”
Breast cancer is one of the leading causes of cancer death worldwide, especially in women. However, early detection significantly increases the success of treatment. For the purpose of early detection, histopathology images should be analyzed accurately. Specifically, during the detection procedure, experts evaluate both general and local tissue organization with whole slide and microscopic images. However, the large amount of data and the complexity of the images make this task time-consuming and cumbersome, with differing interpretations among experts. Therefore, software tools for computer-assisted (automatic) detection need to be developed.
In this study, the definition, preparation and creation stages of NuSeC and MiDeSeC datasets to be used for automatic detection of breast cancer are explained. The stages of creating datasets are shown in Figure 1. In the first stage, all the slide images are divided into patches of 1024×1024 pixels, and in the second stage, the images are labeled/marked on a pixel basis and their masks are created.
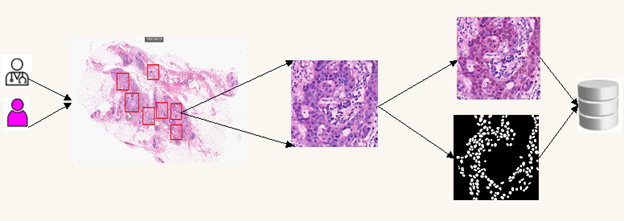
Figure 1: The datasets creation stages
S
The images used in the datasets were created using H&E stained invasive breast carcinoma, special type (NST) slides of 25 different patients produced at Medical Pathology Department of Ankara University with 40x magnification. Slides were scanned with a 3D Histech Panoramic p250 Flash-3 scanner and Olympus BX50 microscope. QuPath software was used to create the datasets.
The datasets were created by the Non-Pathologist (NP) and Pathologist (P) researchers listed below (http://compseg.ankara.edu.tr/arastirmacilar):
1) Prof.Dr. Refik Samet (NP), Ankara University Computer Engineering Department, Computer Engineer;
2) Prof.Dr. Serpil Sak (P), Ankara University Department of Medical Pathology, Pathologist;
3) Assoc.Prof.Dr. Emrah Hancer (NP), Burdur Mehmet Akif Ersoy University, Department of Software Engineering, Computer and Software Engineer;
4) Assoc.Prof.Dr. Bilge Ayça Kırmızı (P), Ankara University, Department of Medical Pathology, Pathologist;
5) Ph.D. Student Zeynep Yıldırım (NP), Ankara University Computer Engineering Department, Computer Engineer;
6) Ph.D. Student Nuşin Nemati (NP), Ankara University Computer Engineering Department, Computer Engineer;
7) Graduate Student Mohamed Traoré (NP), Ankara University Computer Engineering Department, Computer Engineer.
The full slide images used in the datasets were provided by Ankara University Medical Pathology Department. First, all slide images were processed by NP researchers and drafts of the datasets were created. Then, the drafts of the created datasets were checked, corrected and approved by the P researchers of Ankara University Medical Pathology Department. Finally, on the base of the results of the experiments carried out using deep learning models developed within the scope of the project and known in the literature, necessary corrections and changes were made and the final versions of the datasets were created and shared on our website (http://compseg.ankara.edu.tr/en/datasets/).
2. NuSeC – Nuclei Segmentation and Classification Dataset
The NuSeC dataset consists of a total of 100 images. In other words, NuSeC includes four 1024×1024 pixel images from each of the 25 full slide images belonging to 25 patients. NuSeC was divided into two sub-datasets as 75% (75 units) for training and 25% (25 units) for testing purposes.
The test sub-dataset was generated by randomly selecting one of the 4 images produced from each of the 25 full slide images. The training sub-dataset, on the other hand, consists of rest 3 images generated from each of the 25 full slide images. There are approximately 30000 nuclei structures in 75 images in the training sub-dataset. On the other hand, there are approximately 6000 nuclei structures in 25 images in the test sub-dataset. Figure 2 shows sample images of the NuSeC dataset.
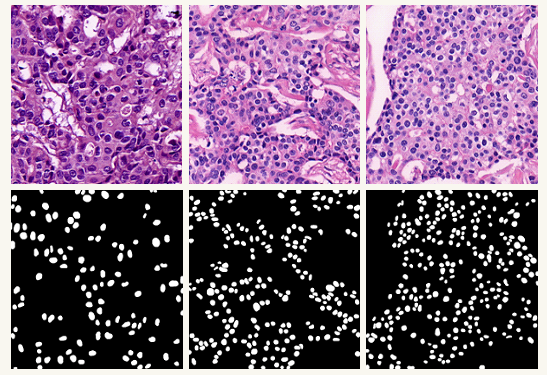
Figure 2: The Sample Images of NuSeC Dataset
S
3.MiDeSeC – Mitosis Detection, Segmentation and Classification Dataset
While creating the MiDeSeC dataset, care was taken to include images of different patients to represent different mitosis patterns. For this purpose, 50 regions with mitotic structures were marked in the full slide images of 25 patients, and 50 images of 1024×1024 pixels were created, one from each of this region. There are approximately 500 mitoses in the images produced. MiDeSeC dataset was created as training and test sub-datasets, 70% (35 images) for training purposes and 30% (15 images) for testing. The test sub-dataset was created by randomly selecting 15 of 50 images of 25 patients. The training sub-dataset, on the other hand, was composed of 35 images, excluding 15 randomly selected ones for the test sub-dataset. Finally, a CSV file with mitosis coordinates was produced. Figure 3 shows sample images of the MiDeSeC data set.
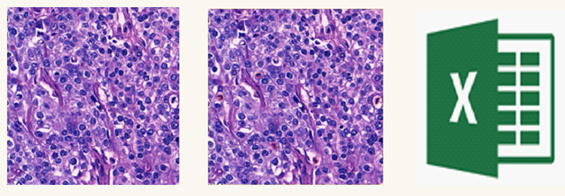
Figure 3: The Sample Images of MiDeSeC Dataset
S
4.Dataset Links
NuSeC Dateset link (http://compseg.ankara.edu.tr/en/datasets/)
MiDeSeC Dataset link (http://compseg.ankara.edu.tr/en/datasets/)
E-mail address for comments and suggestions: compseg@ankara.edu.tr